
One of our BEAR Champions, Dr Jason Turner, described how he makes use of the many different BEAR services to enable his research at a recent seminar for the Institute for Inflammation and Ageing, which BEAR were invited to present at. Read on to find out how quickly he can transfer 260GB of data!
Jason is a post-doc within the Institute of Inflammation and Ageing and whilst he started off as a ‘wet lab’ biologist, he now spends a lot of time on the computational side, looking at single cell RNA sequencing, which he says…
“…requires specific tools to do the analysis and specific ways to store the data and process it, which wouldn’t be possible without the support of the BEAR infrastructure behind it.”
Jason uses many different BEAR services throughout his research involving single-cell sequencing.
Big Data Transfer
Jason receives data from Genomics Birmingham, which previously had to come via ftp servers and data transfer was very slow. Recently they have started using Globus and with BEAR now providing the BEAR Data Transfer service using Globus, it means Jason can get the data directly to the BEAR Research Data Store much more quickly.
“Globus makes it wonderful easy, not only to transfer the data, but it also makes it incredibly rapid to transfer it direct to an RDS drive with not too much complication, it’s a graphical user interface.”
To give an idea of the speed of data transfer, Jason was able to transfer a 30GB file in 1 minute 30 seconds. A 260GB file transferred in just 7 minutes – speeds that Jason is not able to comprehend, allowing him to get the data needed for analysis quickly and securely. Each transfer is encrypted and the file integrity is checked too (see below output).

Data Storage
Jason has 9 core projects that he is actively working on, for which he has access to 27TB of free storage via the BEAR Research Data Store. Jason has also recently purchased 100TB of storage to meet the needs of upcoming projects. He stores multiple 10x genomic data sets, bulk RNA-sequencing datasets, ultrasound images as well as general wet lab data and documents. The project PI or Data Manager controls access to allow others to view and edit data and it integrates with the supercomputer BlueBEAR for High Performance Computing.
Data Analysis
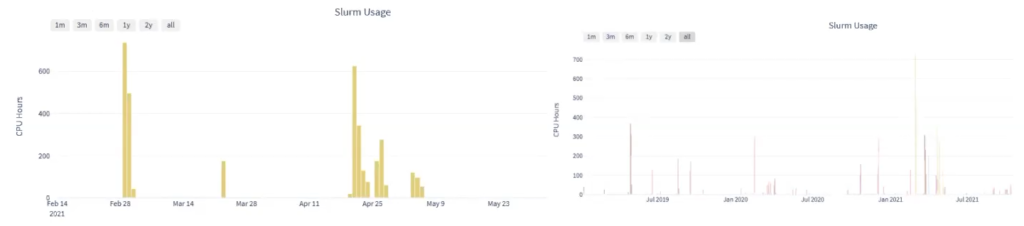
There are an “impressive” amount of applications available to run on the BlueBEAR supercomputer, if the application you need is not on there already there is a simple process to request it to be added. Jason uses thousands of CPU hours across the various projects he is involved in for data processing (one example below shows 700 CPU hours in use), – his laptop only has 8 cores so can only perform 8 CPU hours and to use it for all the data processing would “take a significant amount of time” and would prevent him from using the machine for anything else.
“But actually this is thousands, possibly 10s of thousands of CPU hours, that we wouldn’t be able to work with, we wouldn’t have access to this data essentially without the BlueBEAR infrastructure behind it, so the High Performance Computing for this kind of analysis is just critical.”
Coaching
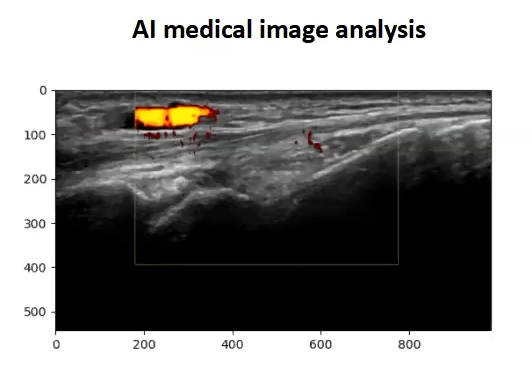
Jason has also made use of the coaching from the Research Software Group. Research Software Engineer (RSE) Simon Hartley has been working with Jason to use AI techniques for medical image analysis, specifically looking at neural network analysis of ultrasound images. Coaching from Simon has enabled Jason to implement automatic grading of ultrasonic images (dicom images), converting the medical grade images to jpegs and tifs, which are more typically used for AI and pulling down the metadata needed to classify the images.
Training
Jason has also made use of the various training courses available from BEAR such as Introduction to Linux, Introduction to BlueBEAR, Software Carpentries on Python and C++ – an almost “endless breadth” of training on offer, and now Jason helps the BEAR team by teaching on the R Software Carpentries workshop after attending Software Carpentries Instructor training, funded by BEAR.
Conclusion
Jason concluded by saying how essential for him and his projects the BEAR infrastructure is and for analysis of the big data sets involved in single cell sequencing.
We were so pleased to hear of how Jason is able to make use of what is on offer from Advanced Research Computing, particularly to hear examples of how quick the data transfer is via Globus – if you have any examples of how it has helped your research then do get in contact with us at bearinfo@contacts.bham.ac.uk. We are always looking for good examples of use of High Performance Computing to nominate for HPC Wire Awards – see our recent winners for more details.
