In this case study, we hear from Xi Jia, who is doing a Ph.D. in the School of Computer Science. Xi has been making use of BEAR’s Baskerville GPU cluster to dramatically reduce the time it takes to train deep-learning models for medical image registration.
I am a fourth-year PhD student, from the School of Computer Science, University of Birmingham, working on deep learning-based medical image registration. My project involves high dimensional volumetric data, such as 3D brain MRI scans, 3D Lung CT scans, and 4D cardiac MRI scans. For these high-resolution medical scans, training a basic 3D deep registration model usually requires at least one GPU (with 40G VRAM) and days to optimize, not to mention the tedious hyperparameters tuning followed. Thanks to the powerful computational resources that Baskerville provided, I was able to train and tune multiple deep registration models in parallel. And our deep registration model trained on Baskerville was the winning solution of the well-known MICCAI 2022 Learn2Reg Challenge -Task 1.
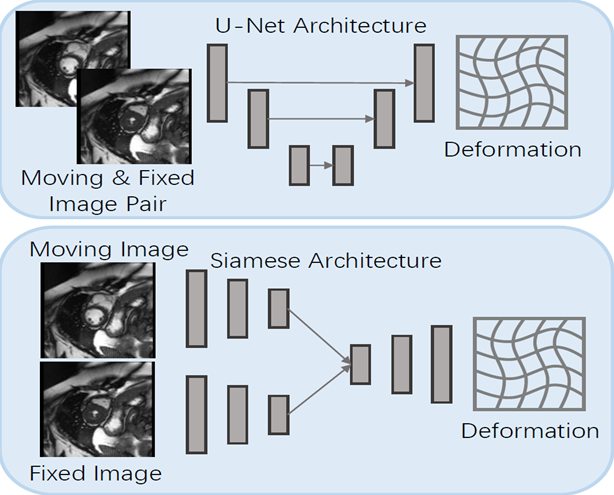
Image registration maps a moving image to a fixed image according to its spatial correspondence. The procedure typically involves two operations: 1) estimating the spatial transformation between the image pair; 2) deforming the moving image with the estimated transformation. In medical image analysis, registration is critical for many automatic analysis tasks such as multi-modality fusion, population modeling, and statistical atlas learning. Deep learning based Registration approaches directly take the moving and fixed images as input and estimate the optimal deformations under a loss criterion. Commonly used network architectures (U-Net and Siamese Network) for deep registration are illustrated in the Figure below. U-Net(-like) approaches concatenate the moving and fixed images into a multi-channel image as input. Siamese Network based approaches take the moving and fixed images separately into the two Siamese branches.
Though deep image registration is conceptively simple, the training of a 3D deep registration model poses a challenge for most research students, as it requires a lot of computational resources. In the table below, using 160x192x220 3D image as an example, I listed the parameters, Multiply-Addition operations, and inference time (one forward propagation in CPU and GPU) of two state-of-the-art registration models.
| Model | Parameters | Mul-Adds (G) | CPU (s) | A100 GPU (s) |
| VoxelMorph | 301,411 | 398.81 | 10.530 | 0.441 |
| TransMorph | 46,771,251 | 657.64 | 22.035 | 0.443 |
As we can see from the table, one single forward propagation of the TransMorph model already takes 657.64 Giga of Multiply-Addition operations and costs about 22 seconds with the CPU. However, training a deep registration model usually need to go through the whole dataset multiple times. If we train a TransMorph model (500 epochs) with a dataset of 400 3D scan pairs, the training time will be an unacceptable 102 days. If we switch to the GPU mode, the training time will be rapidly decreased to about 2 days, which becomes practical.
On the other hand, as deep registration models (such as VoxelMorph and TransMorph) involve lots of Multiply-Addition operations, the training of these models requires sufficient GPU memory. The 40G VRAM of A100 GPU in Baskerville also meets the requirement.
In the last section, we compare the inference time between the CPU and GPU of two SOTA registration models, and we proved the effectiveness of using the Baskerville A100 GPU.
More importantly, there are 4 A100 GPUs in a Baskerville node, which means I am able to train 4 different models in parallel within 2 days. Thanks to the powerful computational resource provided by Baskerville, our extended method of LKU-Net (https://arxiv.org/pdf/2208.04939.pdf ) just won the Task 1 of the MICCAI 2022Learn2Reg Challenge, which is one of the most well-known academic contests in the medical image registration field. I am using and benefiting from Baskerville. Why don’t you give it a try?
We were so pleased to hear of how Xi is able to make use of what is on offer from Advanced Research Computing, particularly to hear of how he has made use of the GPU’s on Baskerville and winning the Task 1 of the MICCAI 2022Learn2Reg Challenge – if you have any examples of how it has helped your research then do get in contact with us at bearinfo@contacts.bham.ac.uk. We are always looking for good examples of use of High Performance Computing to nominate for HPC Wire Awards – see our recent winners for more details.
