In July’s case study we hear from Stanimir Tashev, a PhD student in Cardiovascular Science, who has been making use of BEAR’s storage and data processing power, specifically the BEAR Portal to enable his research…

I have been a Ph.D. student in the Herten Lab (@HertenDirk) since 2019 and my project deals with single molecule microscopy as well as the quantification of protein complexes on the surface of cells. This means that my work includes both a biological component as well as plenty of computational work, requiring both image processing and data management. My research produces many terabytes of data and requires tens of gigabytes of RAM, which is more than a work laptop can handle.
Data Storage
As microscopy can generate a large amount of imaging data in a short time, we require a large and secure storage space. BEAR provides free storage of 3 terabytes (TB) of data per project, which is enough for most research projects and offers the option to purchase much more. For our research, we have multiple subfolders covering data from photobleaching step analysis to single-photon statistic experiments. Now we can generate data without the fear of running low on space.
ImageJ/Fiji
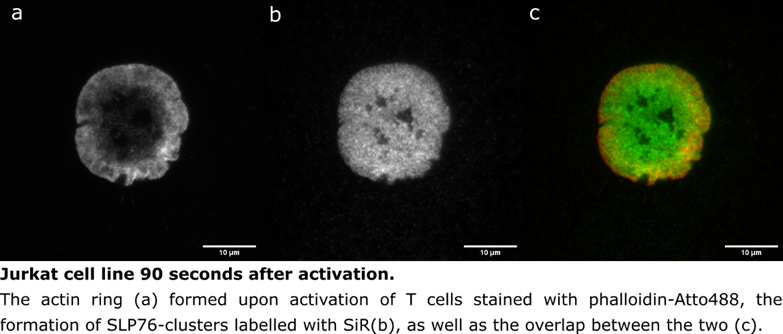
Some of our data processing requires image processing prior to other analyses, which can be done using the software ImageJ and its expansion Fiji. This can include simple things such as setting the correct pixel size and adding a scale bar, or it can be more complex such as running a script that can be modified or analysing a stack of images (see figure below). Fiji can be accessed via the BEAR portal, which also has other interactive apps such as Jupyter Notebooks. Fiji scripts can require different plugins such as ThunderSTORM, which can be easily installed by placing them in an individual’s folder via the Files App. In our case, this lets us start the analysis while the files are safely stored in BEAR.
Python
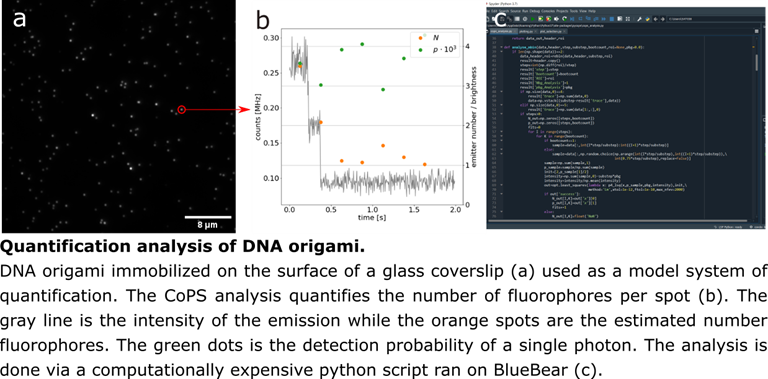
The BlueBEAR supercomputer has a quick and intuitive way to run Python scripts by submitting them as “jobs”. This can be done on data that is already on the Research Data Store, which is accessible by BlueBEAR, and gives us the ability to do computationally heavy analysis on up to 72 cores with 500GB RAM on big data sets. By using BlueBEAR, there is a huge decrease in the time we spend processing data and we are able to perform clustering algorithms to identify the clusters of fluorescence emission through an entire image set, which would be impossible on most computers.
Conclusion
BlueBEAR is suitable for the analysis of microscopy data. It has become the primary way of analysing data for me and is an irreplaceable part of every data processing pipeline of my project, which means that all data passes through BlueBEAR at least once.
We were so pleased to hear of how Stanimir is able to make use of what is on offer from Advanced Research Computing, particularly to hear of how he has made use of the BEAR portal – if you have any examples of how it has helped your research then do get in contact with us at bearinfo@contacts.bham.ac.uk. We are always looking for good examples of use of High Performance Computing to nominate for HPC Wire Awards – see our recent winners for more details.
