In this case study we hear from Thomas Tarenzi, a research fellow in Chemistry, who has been making use of BlueBEAR to enable his research into developing and applying integrative structural biology techniques to biomolecular complexes.

I am a Scientific Officer at HWB-NMR and a member of the team of Professor Teresa Carlomagno. In the group, which includes members from the School of Biosciences, the School of Chemistry, and the Institute of Cancer and Genomic Science, we develop and apply integrative structural biology techniques to biomolecular complexes. Specifically, my research focus lies in the development and application of molecular dynamics techniques for the study of protein-protein and protein-membrane interactions.
Given the size of the systems modelled (of the order of hundreds of thousands of particles) and the complexity of the processes simulated (spanning timescales from nanoseconds to milliseconds), it is fundamental for my job to have access to HPC resources. BlueBEAR fits perfectly to this role.
One of the topics we are particularly interested in is the formation of high-affinity complexes between disordered protein regions. These “fuzzy” protein complexes show neither signs of site-specific interactions nor structure formation, and therefore represent a novelty with respect to the traditional notion that proteins bind to each other through their complementary shapes. Despite being fairly new, the concept that fuzzy interactions can lead to the formation of high-affinity complexes has been demonstrated for a number of cases: one of this is the acetylation mechanism of the histone 3 (H3) tail, a chemical modification that can modulate chromatin structure and affect DNA accessibility in the nucleosome.
it is fundamental for my job to have access to HPC resources. BlueBEAR fits perfectly to this role.
This reaction is assisted by the histone chaperones Asf1 and Vps75, which form a large complex with the acetyltransferase enzyme Rtt109 and the histone dimer H3:H4 (Figure 1). It was recently shown that Vps75 promotes histone acetylation by engaging the disordered H3 N-terminal tail in fuzzy electrostatic interactions with its disordered C-terminal domain, thereby confining the H3 tail to a wide central cavity faced by the Rtt109 active site (Danilenko et al., Nature Communications, 2019). However, the details of how this process takes place are still unclear.
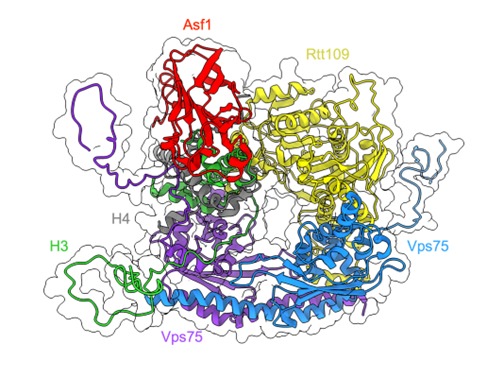
We are currently characterising the fuzzy complex between H3 and Vps75 tails using a combination of unbiased and biased molecular dynamics simulations. In the latter case, we employ a technique called metadynamics, which allows us to accelerate the sampling of rare events and the transitions between conformational states by applying specifically designed biasing potentials on carefully chosen system properties. In this way, we can efficiently identify the interactions between the disordered protein regions, and we can assess if and how the formation of transient structures affects the interaction patterns. These results can help to better understand this important step in the regulation of cellular processes, and at the same time to shed light on an elusive type of bimolecular interaction that challenges the traditional view of Structural Biology.
This process is unthinkable without the availability of large-scale computing resources; BlueBEAR capabilities are therefore crucial to perform the parallel computing required for my research.
Despite the use of such advanced computational techniques, a thorough sampling still requires the acquisition of long trajectories. Making use of an HPC cluster and a highly-scalable molecular dynamics software, simulations of this type can take several weeks or even months to reach convergence. This process is unthinkable without the availability of large-scale computing resources; BlueBEAR capabilities are therefore crucial to perform the parallel computing required for my research.
We were so pleased to hear of how Thomas was able to make use of what is on offer from Advanced Research Computing, particularly to hear of how he has made use of BlueBEAR HPC and its many cores – if you have any examples of how it has helped your research then do get in contact with us at bearinfo@contacts.bham.ac.uk. We are always looking for good examples of use of High Performance Computing to nominate for HPC Wire Awards – see our recent winners for more details.
