In this case study we hear from Sophie Wetz, a Masters student in Psychology, who has been using BlueBEAR to investigate whether a dynamic, rising boundary improved the fit of the full Drift Diffusion Model (DDM) to Random Dot Kinematogram (RDK) task data.

I’m a Master’s student on the Computational Neuroscience stream in the School of Psychology, University of Birmingham. My project tested whether a dynamic, rising boundary improved the fit of the full Drift Diffusion Model (DDM) to Random Dot Kinematogram (RDK) task data, relative to a fixed boundary. Such rising boundaries may emerge in situations when initial information is unreliable or when delaying a response allows additional evidence accumulation.
Methodologically, the work centred on population-based Bayesian inference. We used Differential Evolution Markov Chain Monte Carlo (DE-MCMC) methods because dynamic boundary models produce high-dimensional, correlated parameter spaces where conventional random-walk MCMC mixes poorly. DE-MCMC evolves many chains in parallel and proposes moves using information from other chains, improving mixing and reducing the risk of local optima. Likelihoods were approximated with online Kernel Density Estimation (oKDE), while model fit was assessed with cumulative distribution functions (CDFs), error rates and conditional accuracy functions (CAFs). Alongside this, participant-level simulations based on Expected A Posteriori parameter estimates were examined. We experimented with boundary functions including logistic-sigmoidal, plateaued-linear and exploratory Weibull variants.
The cluster’s high core counts and fast shared storage turned what would have been multi-week, single-machine jobs into multi-day batches.
BlueBEAR made this heavy lifting feasible and reproducible. Running MATLAB R2021b on the BlueBEAR high-performance computing cluster provided computational resources optimised for large-scale parallel computation, ensuring efficient Bayesian estimation and model evaluation. The cluster’s high core counts and fast shared storage turned what would have been multi-week, single-machine jobs into multi-day batches. That faster turnaround meant I could iterate quickly on model variants and priors, tighten convergence criteria, and run more thorough posterior predictive checks rather than compromising for time.
Given the time restraints and complexity of this project, the BlueBEAR’s facilities were essential for my research. Overall, the findings provided partial support for dynamic, rising thresholds in perceptual decisions and highlighted the need for more flexible boundary forms and formal model-comparison criteria. Future work will extend Weibull-shaped boundaries and add such Bayes factor/BIC-based comparisons, where BlueBEAR will again be central to making those analyses possible.
Note. These diagrams are provided for visual context only and do not exactly represent the finalised fitted models
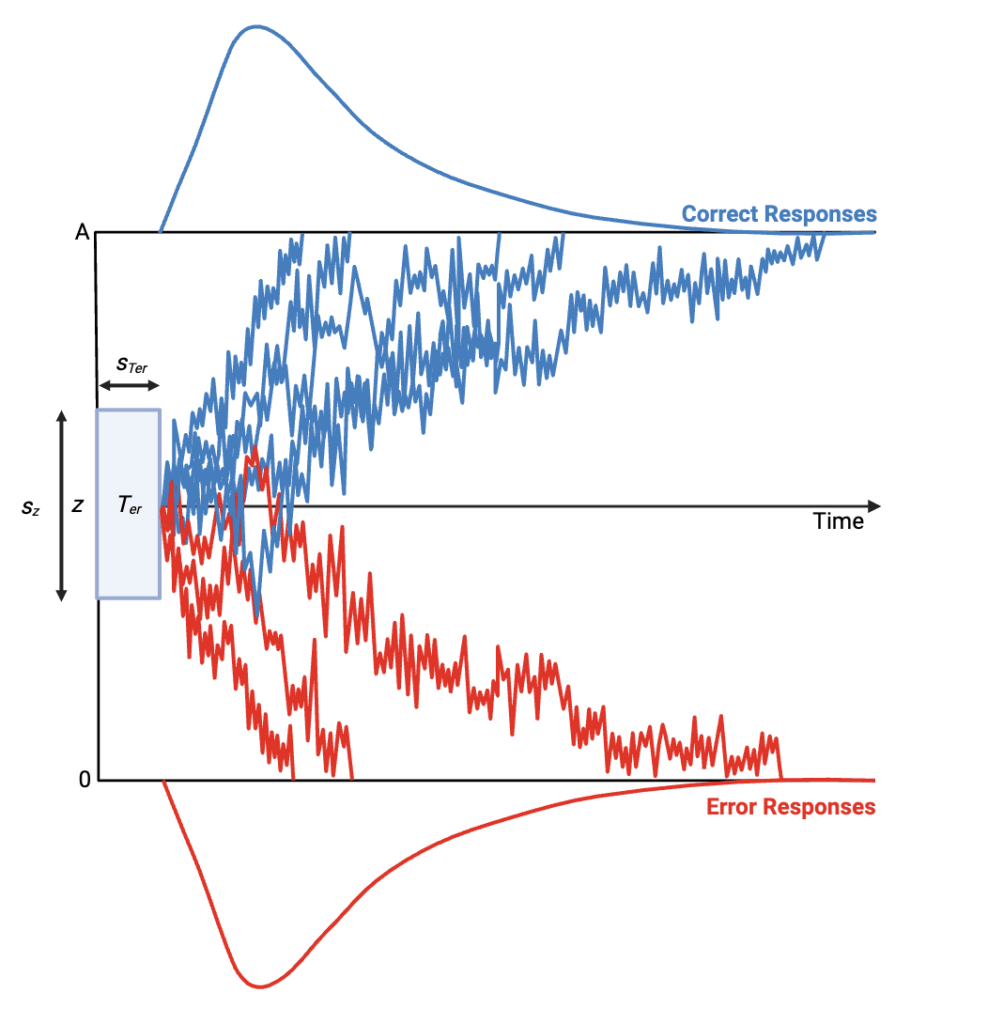
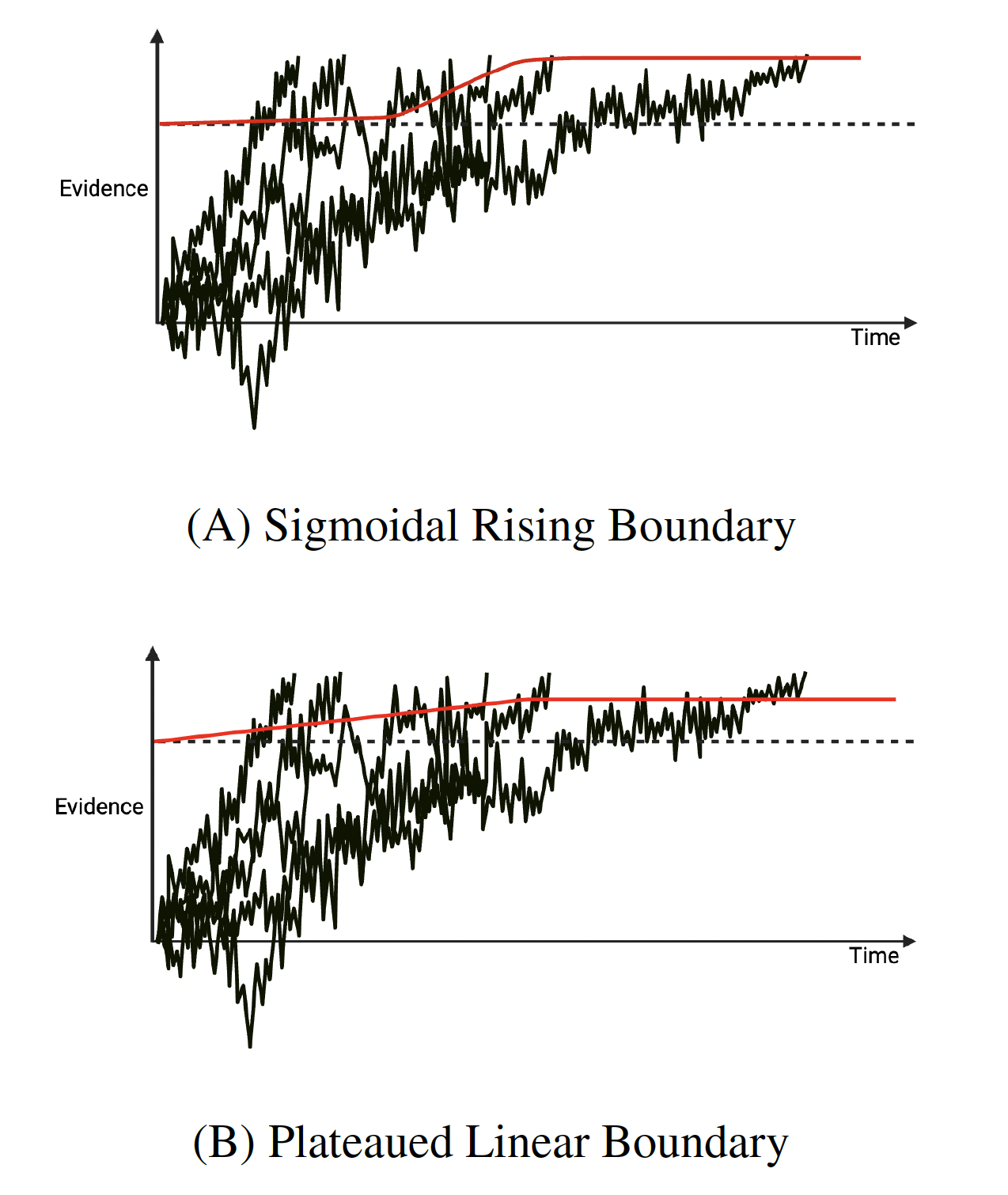
We were so pleased to hear of how Sophie was able to make use of what is on offer from Advanced Research Computing, particularly to hear of how they have made use of the BEAR compute and storage, – if you have any examples of how it has helped your research then do get in contact with us at bearinfo@contacts.bham.ac.uk.
We are always looking for good examples of use of High Performance Computing to nominate for HPC Wire Awards – see our recent winner for more details.
