In this case study, we hear from Samuel Dettmer (Chemistry), who has been making use of BlueBEAR and the Research Data Store to enable his research into the ability of metal compounds to bind to DNA structures.

My name’s Sam and I’m a 2nd year PhD student in the Hannon Group in the School of Chemistry. My research project investigates the ability of metal compounds to bind to DNA structures. This is an important area of medicinal chemistry in which we are trying to understand if we can target specific DNA structures that occur during particular cell processes, like DNA replication for example, and achieve a therapeutic effect. We are currently interested particularly in antiviral activity (against Sars-CoV-2, which causes COVID-19) and anticancer activity.
Whilst much of my research relies on analytical lab work to elucidate interactions between my compounds, a deeper understanding of what is happening at the atomic level can be obtained by using molecular dynamics simulations. These simulations allow me to visualise the movement of DNA and its interactions with water, salt and my compounds over a short timescale (up to 10 microseconds typically). Typically in drug discovery, chemists more frequently search for small (non-metallic) compounds that can target proteins.
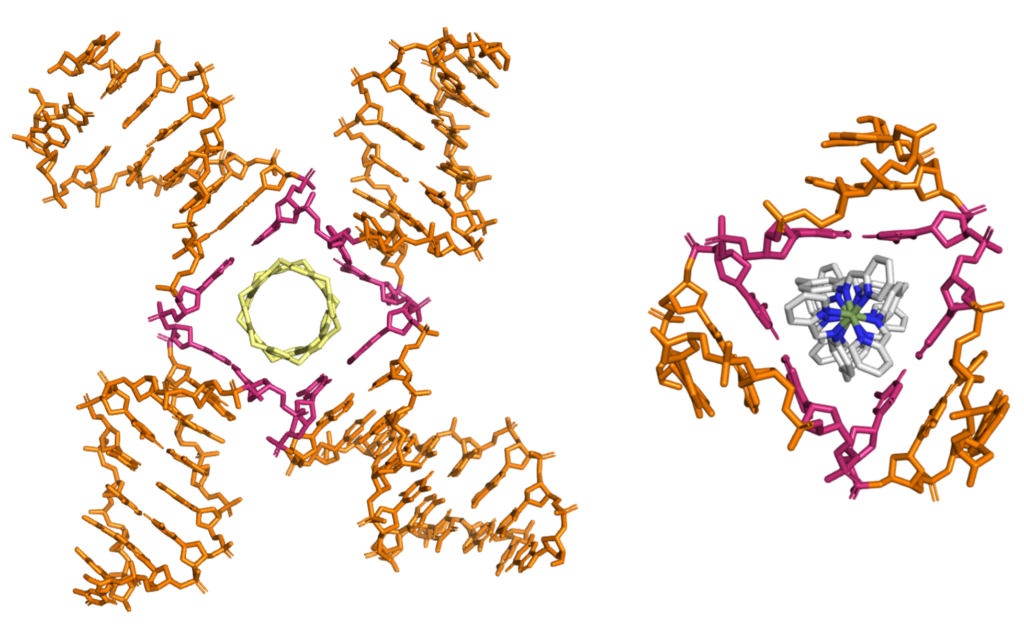
(Craig, J. S.; Melidis, L.; Williams, H. D.; Dettmer, S. J.; Heidecker, A. A.; Altmann, P. J.; Guan, S.; Campbell, C.; Browning, D. F.; Sigel, R. K. O.; Johannsen, S.; Egan, R. T.; Aikman, B.; Casini, A.; Pöthig, A.; Hannon, M. J., Organometallic Pillarplexes That Bind DNA 4-Way Holliday Junctions and Forks. J. Am. Chem. Soc 2023. https://doi.org/10.1021/jacs.3c00118)
A computational method used for this is protein docking, in which the protein is treated as a rigid structure (i.e. it cannot move or rotate) and the computer tries to find the best places that the drug of interest can fit into the protein structure and bind. This approach would simply not work in my case as DNA is a very flexible biomolecule, meaning that it can adopt multiple different shapes and structures that my drug may interact with. Molecular dynamics simulations remove rigidity and allow the DNA atoms to move and rotate freely and ultimately produce a video that I can watch of my DNA moving around and interacting with my compounds.
So where does BEAR come into this? Well, a standard molecular dynamics simulation that I would run of 1 microsecond length typically involves calculating the 3-dimensional movement (and associated energy) of around 50 thousand atoms for 500 million steps! That’s a lot of maths that the computer needs to do and it is simply not possible on your average laptop – it would take weeks if not longer to complete one simulation. In addition to this, the output file of that simulation would be around 80 GB, which is about 15% of my laptop’s total storage capacity – I would quickly fill up my laptop storage with simulation data. BEAR offers me the resources I need to run up to 5 simulations simultaneously on 100 CPU cores each and store all the data long term. Even with these resources, simulations still take up to 4 days to complete, which goes to show just how unreasonably long it would take without the high performance resources.
BEAR offers me the resources I need to run up to 5 simulations simultaneously on 100 CPU cores each and store all the data long term
We were so pleased to hear of how Sam is able to make use of what is on offer from Advanced Research Computing, particularly to hear of how he has made use of BlueBEAR HPC – if you have any examples of how it has helped your research then do get in contact with us at bearinfo@contacts.bham.ac.uk. We are always looking for good examples of use of High Performance Computing to nominate for HPC Wire Awards – see our recent winners for more details.
